The main goals of our research program are to understand how evolutionary processes have shaped human genomes and to quantify the functional relevance of specific genetic differences between individual humans and closely related species. Achieving these goals is critically important to answering fundamental questions about human evolution and to the application of genomics in clinical settings.
We address these questions by applying algorithms and statistical models to ancient and modern genomes. We integrate large-scale genome-wide genotype and phenotype data, including electronic health record data. We also collaborate closely with clinicians and disease-focused biologists, in particular in the Undiagnosed Diseases Network and the March of Dimes Prematurity Research Center.
We have three specific research foci. The highlighted papers illustrate the questions we address and methods we use in the lab. A full list of publications is here.
Modeling the genetics of recent human evolution and disease

- Neanderthal introgression reintroduced functional alleles lost in the human out of Africa bottleneck. DC Rinker, C Simonti, E McArthur, D Shaw, E Hodges, and JA Capra. Nature Ecology & Evolution, 4:1332–1341, 2020. [Paper]
- Inferred divergent gene regulation in archaic hominins reveals potential phenotypic differences. LL Colbran, ER Gamazon, D Zhou, P Evans, NJ Cox, and JA Capra. Nature Ecology & Evolution 3, 1598–1606 (2019). [Paper] [Highlight]
- The phenotypic legacy of admixture between modern humans and Neanderthals. CN Simonti, B Vernot, L Bastarache, …, WS Bush, JM Akey, JC Denny, and JA Capra. Science, 351 (6274):737-741, 2016. [Paper]
Using machine learning to interpret gene regulatory “programs” encoded in genomes
- Learning and interpreting the gene regulatory grammar in a deep learning framework. Chen L and Capra JA. PLOS Computational Biology, 16(11): e1008334, 2020. [Paper]
- Sequence Characteristics Distinguish Transcribed Enhancers from Promoters and Predict Their Breadth of Activity. LL Colbran, L Chen, and JA Capra. Genetics, 301895, 2019. [Paper]
- Prediction of gene regulatory enhancers across species reveals evolutionarily conserved sequence properties. L Chen, AE Fish, and JA Capra. PLOS Computational Biology, 14(10): e100648, 2018. [Paper]
Integrating population genetics and structural biology to enable personalized exome interpretation
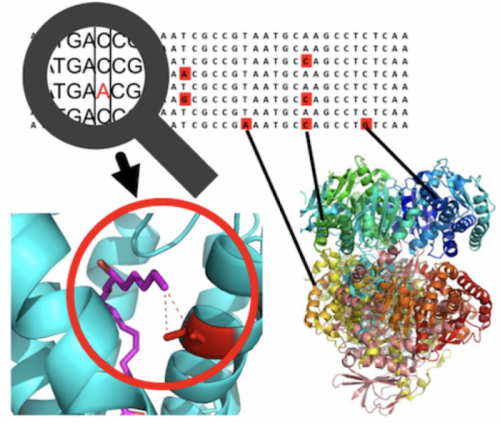
- PSCAN: Spatial scan tests guided by protein structures improve complex disease gene discovery and signal variant detection. Tang, Z. Z., Sliwoski, G. R., Chen, G., Jin, B., Bush, W. S., Li, B., and Capra, J. A. Genome Biology, 21(1), 1-25. [Paper]
- Comprehensive Analysis of Constraint on the Spatial Distribution of Missense Variants in Human Protein Structures. Sivley RM, Dou X, Meiler J, Bush WS*, and Capra JA*. American Journal of Human Genetics, 102:415–426, 2018. [Paper]
- Three-dimensional spatial analysis of missense variants in RTEL1 identifies pathogenic variants in patients with Familial Interstitial Pneumonia. Sivley RM, Sheehan JH, Kropski JA, Cogan J, Blackwell TS, Phillips JA, Bush WS, Meiler J and Capra JA*. BMC Bioinformatics, 19:18, 2018. [Paper]
We are grateful to the NIH, the March of Dimes, and the Burroughs Wellcome Fund for their support.